Estimates the annual phenological cycle from a time series of vegetation greenness.
Arguments
- x
Numeric vector. A time series of a vegetation index (e.g. LAI, NDVI, EVI) or any other variable with seasonal behavior. The code has been optimized to work with integer values. Please re-scale the input values if necessary (e.g. NDVI ranging from 0.0000 to 1.0000, multiply by 10,000).
- dates
A date vector. The number of dates must be equal to the number of "x" values (numeric input vector).
- h
Numeric. Indicates the geographic hemisphere to define the starting date of the growing season. h = 1 if the vegetation is in the Northern Hemisphere (season starting at January 1st), h = 2 if it is in the Southern Hemisphere (season starting at July 1st)
- frequency
Character string. Defines the number of samples for the output phenology and must be one of the following options: 'daily' giving output vector of length 365, '8-days' giving output vector of length 46 (i.e MOD13Q1 and MYD13Q1), 'monthly' giving output vector of length 12,'bi-weekly' giving output vector of length 24 (i.e. GIMMS) or '16-days' (default) giving output vector of length 23 (i.e MOD13Q1 or MYD13Q1).
- rge
Numeric vector with two values setting the minimum and maximum values of the response variable (e.g. NDVI) used in the analysis. We suggest the use of theoretically based limits. For example in the case of MODIS NDVI or EVI, it ranges from 0 to 10,000, so rge = c(0,10000)
- plot
Logical. Set TRUE (default) or FALSE to show the phenology curve plot.
Details
Derives the annual phenological cycle for a standard growing season using a numeric vector of vegetation canopy greenness values (e.g. Leaf Area Index, LAI) or satellite based greenness proxies such as the Normalized Difference Vegetation Index (NDVI) or Enhanced Vegetation Index (EVI). A vector with dates for the greenness values is also required.
Examples
 # \donttest{
library(lubridate)
library(terra)
## Testing raster data from Central Chile (NDVI), h=2##
# Load data
f <- system.file("extdata/MegaDrought_spatRast.rda", package = "npphen")
MegaDrought <- readRDS(f)
# Dates
data("modis_dates")
# Generate a Raster time series using a raster stack and a date database from Central Chile
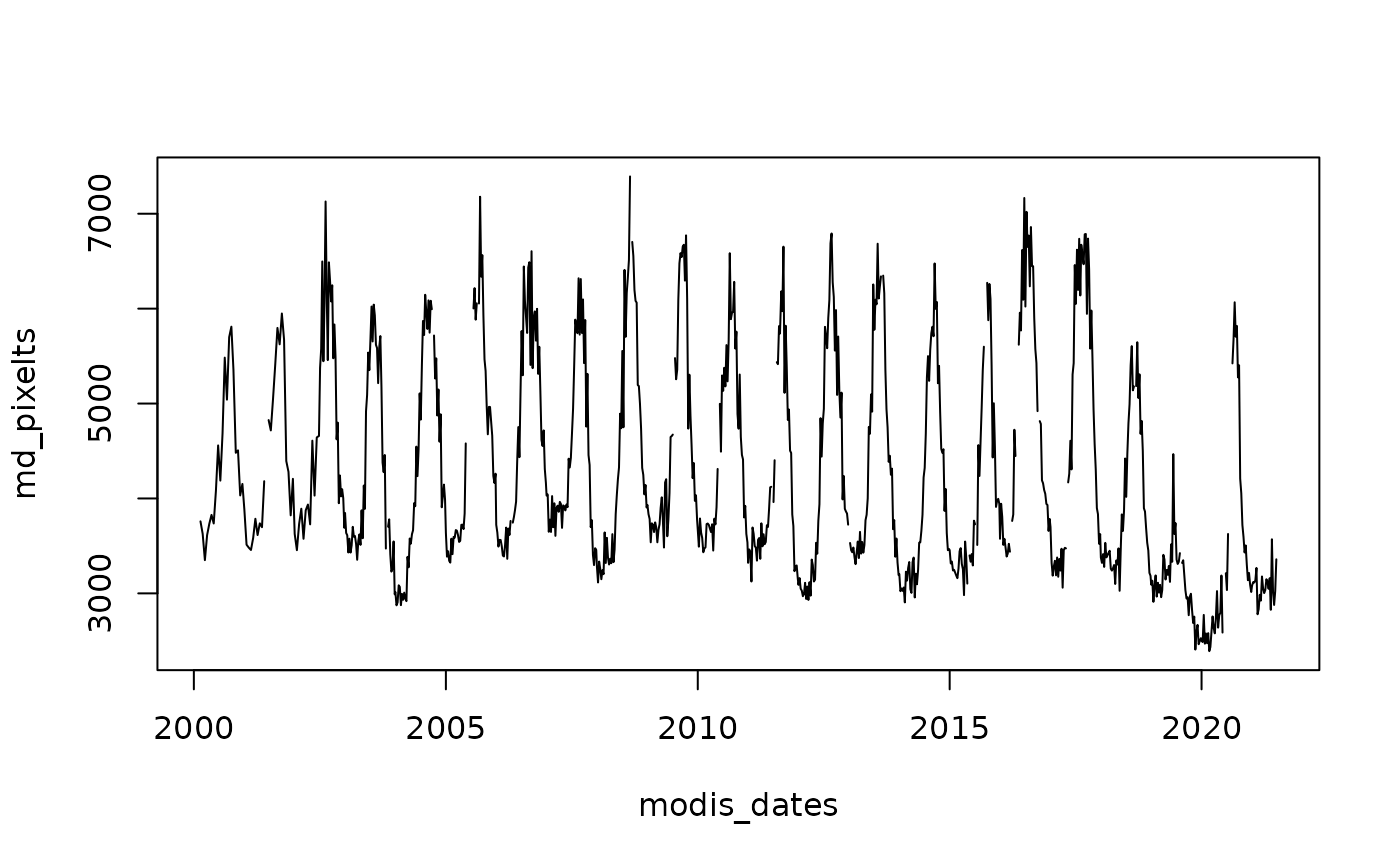
# Obtain data from a particular pixel generating a time series
md_pixel <- cellFromXY(MegaDrought, cbind(313395, 6356610))
md_pixelts <- as.numeric(MegaDrought[md_pixel])
plot(modis_dates, md_pixelts, type = "l")
# \donttest{
library(lubridate)
library(terra)
## Testing raster data from Central Chile (NDVI), h=2##
# Load data
f <- system.file("extdata/MegaDrought_spatRast.rda", package = "npphen")
MegaDrought <- readRDS(f)
# Dates
data("modis_dates")
# Generate a Raster time series using a raster stack and a date database from Central Chile
# Obtain data from a particular pixel generating a time series
md_pixel <- cellFromXY(MegaDrought, cbind(313395, 6356610))
md_pixelts <- as.numeric(MegaDrought[md_pixel])
plot(modis_dates, md_pixelts, type = "l")
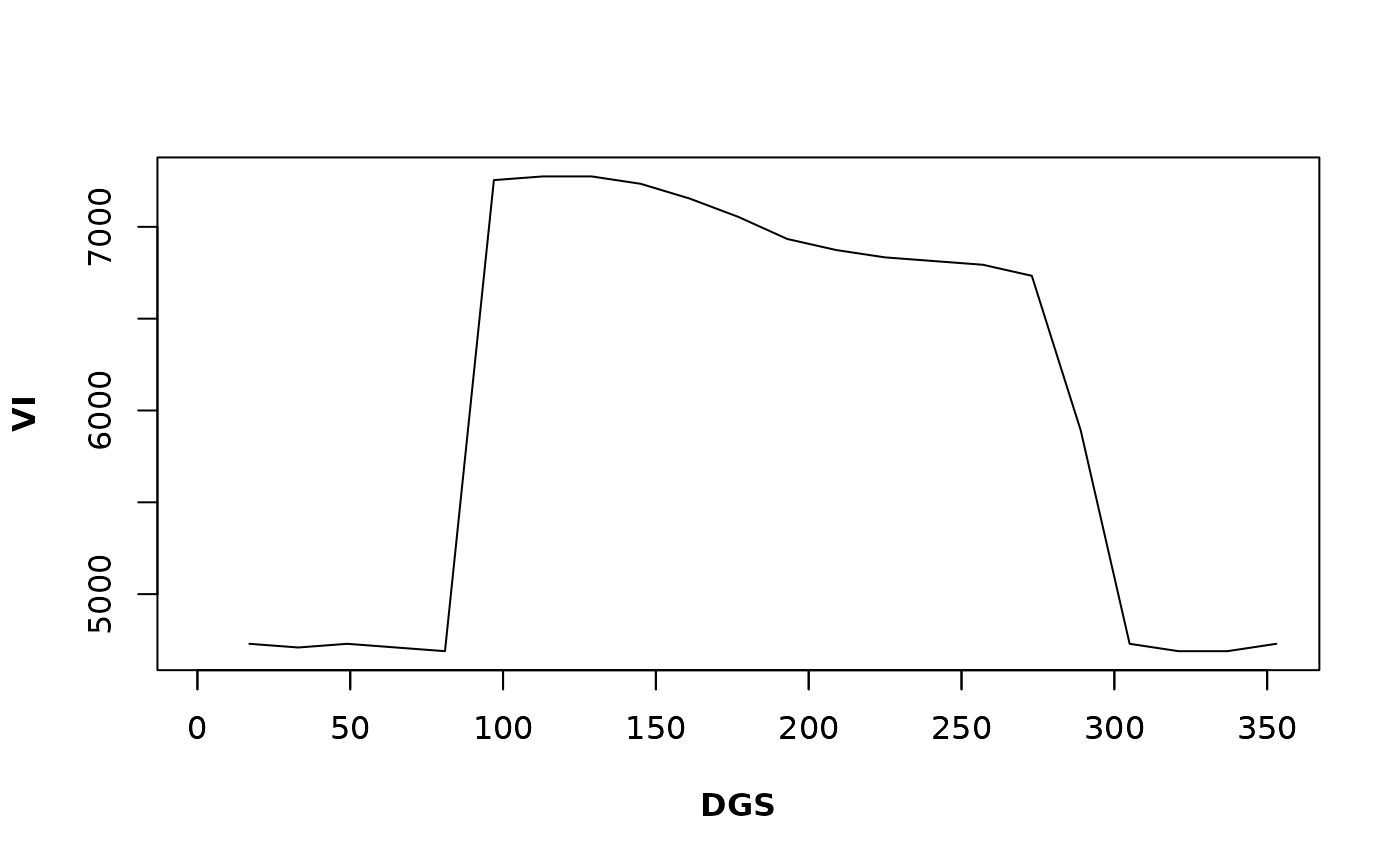
 # Phenology for the given pixel
Phen(x = md_pixelts, dates = modis_dates, h = 2, frequency = "16-days", rge = c(0, 10000))
# Phenology for the given pixel
Phen(x = md_pixelts, dates = modis_dates, h = 2, frequency = "16-days", rge = c(0, 10000))
 #> 1 17 33 49 65 81 97 113
#> NA 5771.543 5871.743 5831.663 5811.623 5911.824 5871.743 4789.579
#> 129 145 161 177 193 209 225 241
#> 4549.098 4148.297 3907.816 3687.375 3486.974 3426.854 3426.854 3446.894
#> 257 273 289 305 321 337 353
#> 3466.934 3466.934 3507.014 3567.134 3667.335 3767.535 3787.575
## Testing with the Bdesert_stack from the Atacama Desert, Northern Chile (NDVI), h=2 ##
# Load data
# SparRaster
f <- system.file("extdata/Bdesert_spatRast.rda", package = "npphen")
Bdesert <- readRDS(f)
# Generate a Raster time series using a raster stack and a date database from Northern Chile
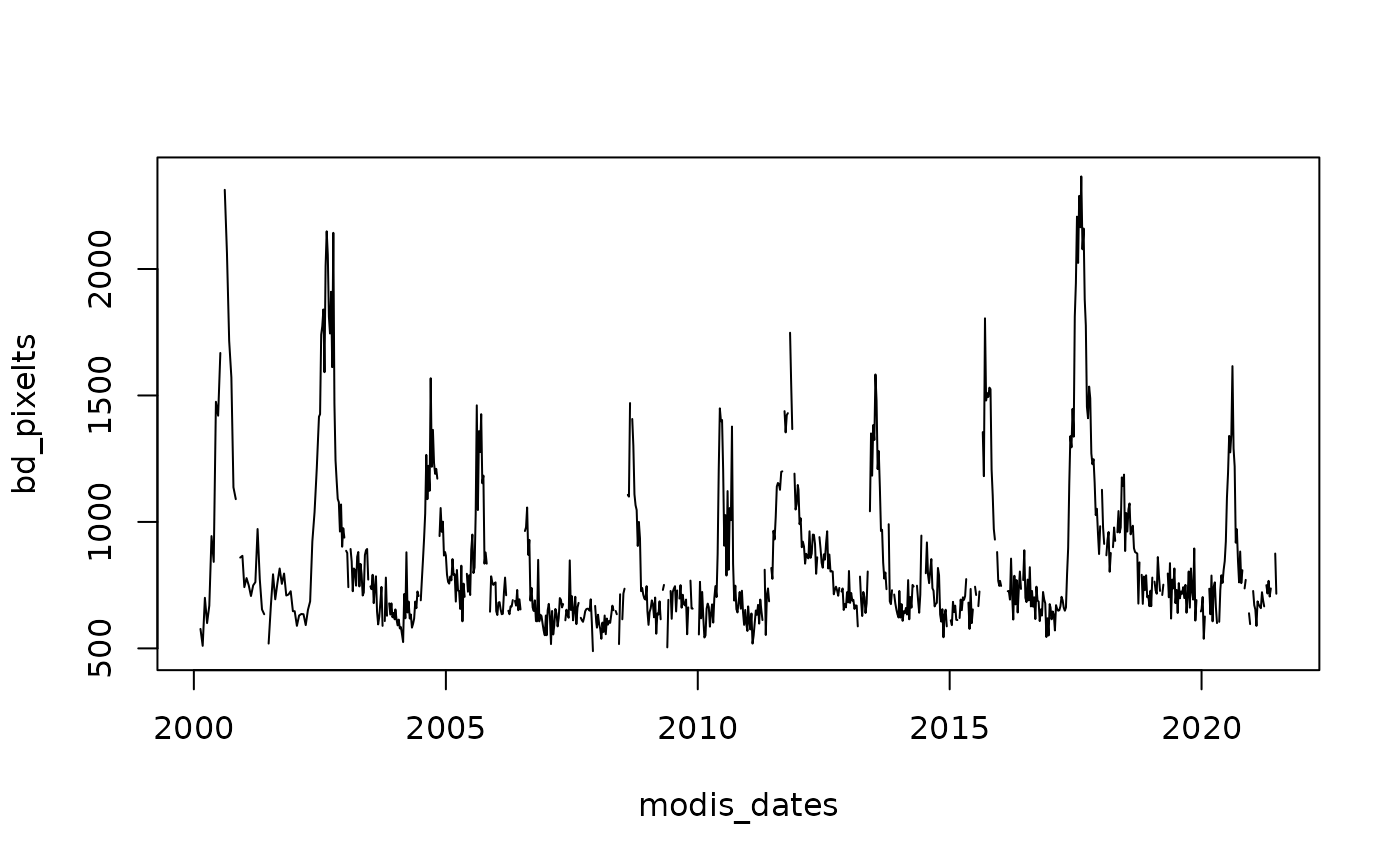
# Obtain data from a particular pixel generating a time series
bd_pixel <- cellFromXY(Bdesert, cbind(286638, 6852107))
bd_pixelts <- as.numeric(Bdesert[bd_pixel])
plot(modis_dates, bd_pixelts, type = "l")
#> 1 17 33 49 65 81 97 113
#> NA 5771.543 5871.743 5831.663 5811.623 5911.824 5871.743 4789.579
#> 129 145 161 177 193 209 225 241
#> 4549.098 4148.297 3907.816 3687.375 3486.974 3426.854 3426.854 3446.894
#> 257 273 289 305 321 337 353
#> 3466.934 3466.934 3507.014 3567.134 3667.335 3767.535 3787.575
## Testing with the Bdesert_stack from the Atacama Desert, Northern Chile (NDVI), h=2 ##
# Load data
# SparRaster
f <- system.file("extdata/Bdesert_spatRast.rda", package = "npphen")
Bdesert <- readRDS(f)
# Generate a Raster time series using a raster stack and a date database from Northern Chile
# Obtain data from a particular pixel generating a time series
bd_pixel <- cellFromXY(Bdesert, cbind(286638, 6852107))
bd_pixelts <- as.numeric(Bdesert[bd_pixel])
plot(modis_dates, bd_pixelts, type = "l")
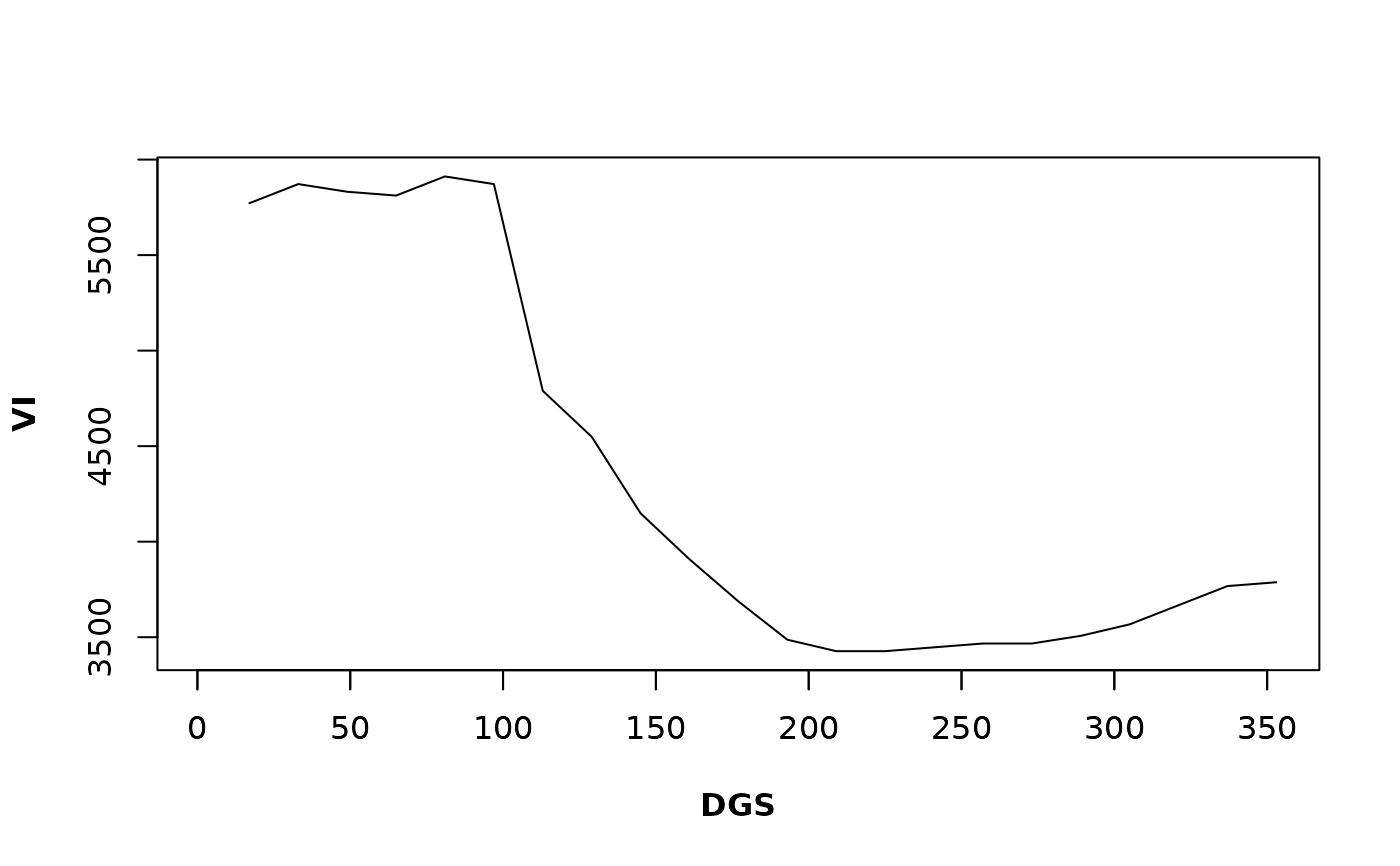
 # Phenology for the given pixel
Phen(x = bd_pixelts, dates = modis_dates, h = 2, frequency = "16-days", rge = c(0, 10000))
# Phenology for the given pixel
Phen(x = bd_pixelts, dates = modis_dates, h = 2, frequency = "16-days", rge = c(0, 10000))
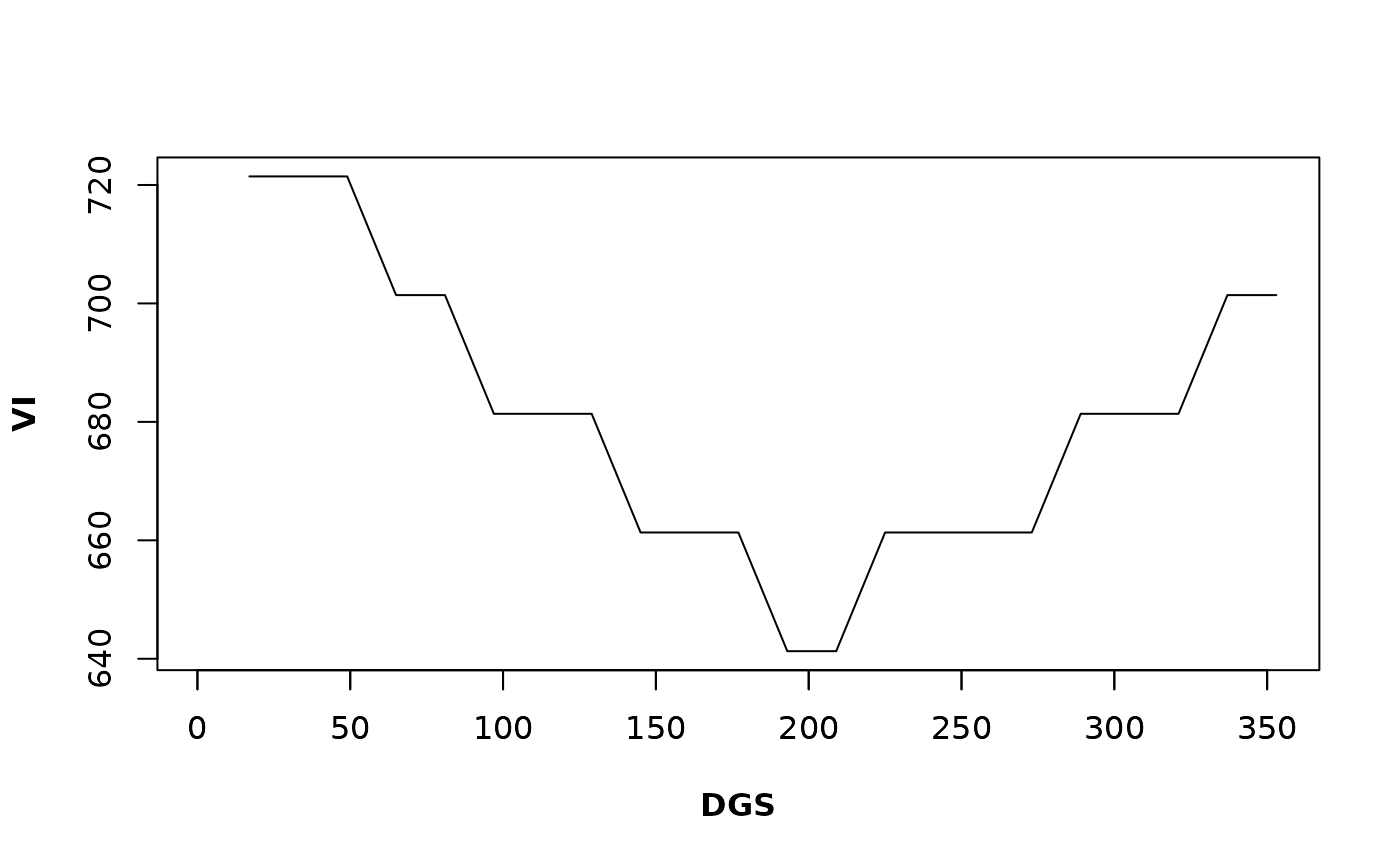 #> 1 17 33 49 65 81 97 113
#> NA 721.4429 721.4429 721.4429 701.4028 701.4028 681.3627 681.3627
#> 129 145 161 177 193 209 225 241
#> 681.3627 661.3226 661.3226 661.3226 641.2826 641.2826 661.3226 661.3226
#> 257 273 289 305 321 337 353
#> 661.3226 661.3226 681.3627 681.3627 681.3627 701.4028 701.4028
# }
#> 1 17 33 49 65 81 97 113
#> NA 721.4429 721.4429 721.4429 701.4028 701.4028 681.3627 681.3627
#> 129 145 161 177 193 209 225 241
#> 681.3627 661.3226 661.3226 661.3226 641.2826 641.2826 661.3226 661.3226
#> 257 273 289 305 321 337 353
#> 661.3226 661.3226 681.3627 681.3627 681.3627 701.4028 701.4028
# }